DREAMS: deep read-level error model for sequencing data applied to
Por um escritor misterioso
Last updated 11 junho 2024
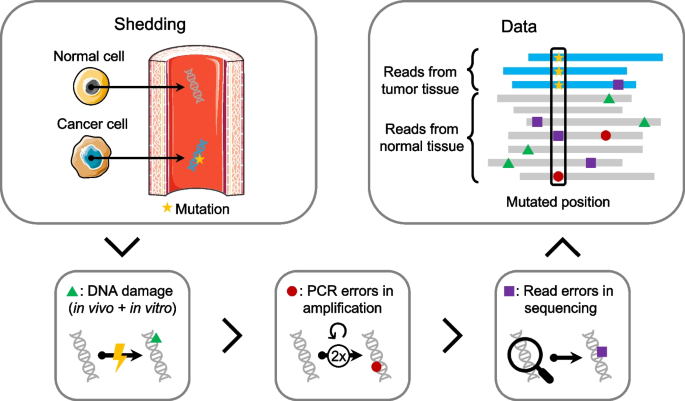
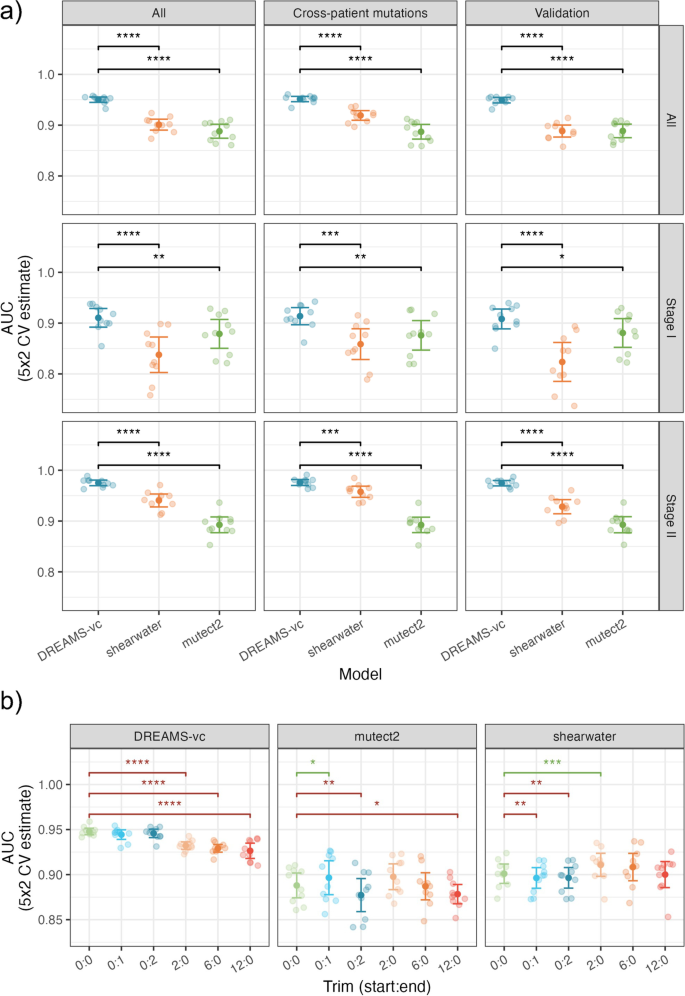
Circulating tumor DNA detection using next-generation sequencing (NGS) data of plasma DNA is promising for cancer identification and characterization. However, the tumor signal in the blood is often low and difficult to distinguish from errors. We present DREAMS (Deep Read-level Modelling of Sequencing-errors) for estimating error rates of individual read positions. Using DREAMS, we develop statistical methods for variant calling (DREAMS-vc) and cancer detection (DREAMS-cc). For evaluation, we generate deep targeted NGS data of matching tumor and plasma DNA from 85 colorectal cancer patients. The DREAMS approach performs better than state-of-the-art methods for variant calling and cancer detection.

What is Text Analysis? A Beginner's Guide
DREAMS: deep read-level error model for sequencing data applied to

PDF) DREAMS: deep read-level error model for sequencing data

Machine Learning in Predictive Toxicology: Recent Applications and

Stop Stealing Dreams. if you don't underestimate me, I…
Life, Free Full-Text
DeSP: a systematic DNA storage error simulation pipeline

SpikingJelly: An open-source machine learning infrastructure

How Dare They Peep into My Private Life?”: Children's Rights
Recomendado para você
-
 The Based God - SCP Foundation11 junho 2024
The Based God - SCP Foundation11 junho 2024 -
SCP Foundation: SCP-7143-J [SFW?]11 junho 2024
-
 US20120297504A1 - Isolated polynucleotides and polypeptides and methods of using same for increasing plant yield, biomass, growth rate, vigor, oil content, abiotic stress tolerance of plants and nitrogen use efficiency - Google Patents11 junho 2024
US20120297504A1 - Isolated polynucleotides and polypeptides and methods of using same for increasing plant yield, biomass, growth rate, vigor, oil content, abiotic stress tolerance of plants and nitrogen use efficiency - Google Patents11 junho 2024 -
 S I M P SCP Foundation (RP) Amino11 junho 2024
S I M P SCP Foundation (RP) Amino11 junho 2024 -
 Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum11 junho 2024
Machimaho: I Messed Up and Made the Wrong Person Into a Magical Girl! - AnimeSuki Forum11 junho 2024 -
 PDF) Public sector productivity: measurement challenges, performance information and prospects for improvement11 junho 2024
PDF) Public sector productivity: measurement challenges, performance information and prospects for improvement11 junho 2024 -
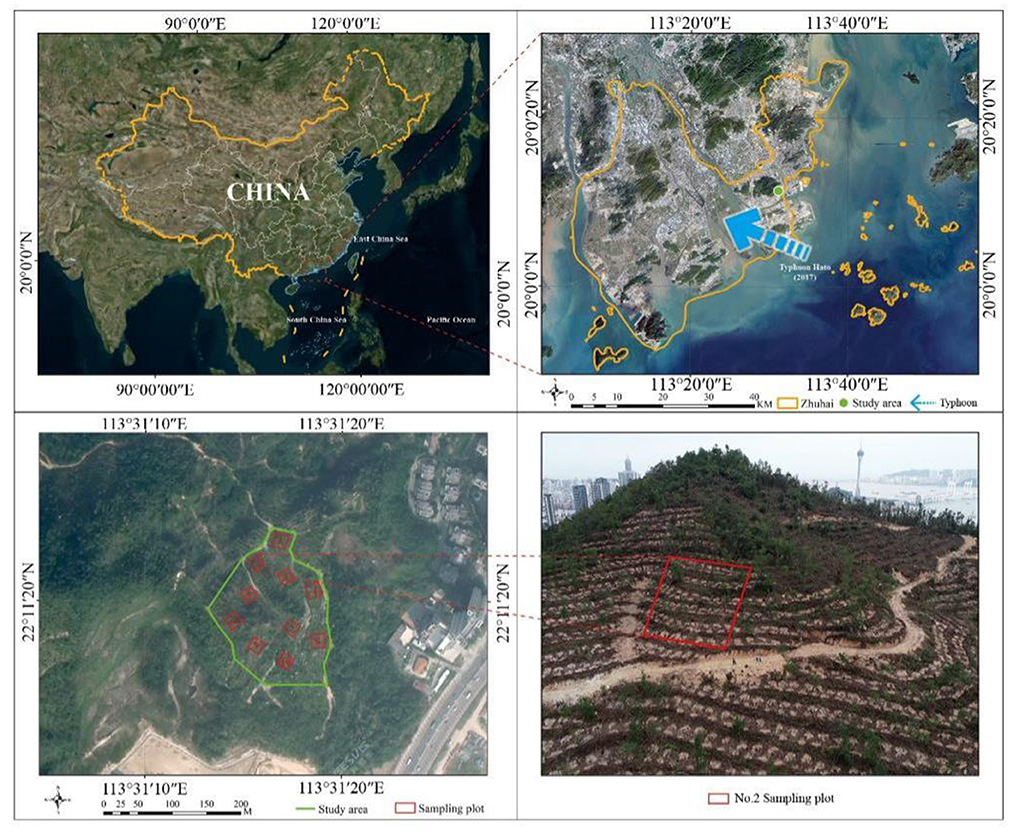 Frontiers Growth patterns and environmental adaptions of the tree species planted for ecological remediation in typhoon-disturbed areas—A case study in Zhuhai, China11 junho 2024
Frontiers Growth patterns and environmental adaptions of the tree species planted for ecological remediation in typhoon-disturbed areas—A case study in Zhuhai, China11 junho 2024 -
 DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology11 junho 2024
DREAMS: deep read-level error model for sequencing data applied to low-frequency variant calling and circulating tumor DNA detection, Genome Biology11 junho 2024 -
 Transgenic Mice Expressing Green Fluorescent Protein under the11 junho 2024
Transgenic Mice Expressing Green Fluorescent Protein under the11 junho 2024 -
 Inhibition of HIV-1 Replication and Activation of RNase L by11 junho 2024
Inhibition of HIV-1 Replication and Activation of RNase L by11 junho 2024
você pode gostar
-
 Coin Master hack: apk mostra como ter dinheiro e giros infinitos no Android11 junho 2024
Coin Master hack: apk mostra como ter dinheiro e giros infinitos no Android11 junho 2024 -
 announces new in-game content, free games for March 202311 junho 2024
announces new in-game content, free games for March 202311 junho 2024 -
 Monkey Mart Unblocked - Play Online Games Free11 junho 2024
Monkey Mart Unblocked - Play Online Games Free11 junho 2024 -
 Sapnap - dsfg11 junho 2024
Sapnap - dsfg11 junho 2024 -
DG-RA Style Chess Pieces Set - Openclipart11 junho 2024
-
 Camiseta Feminina Baby Look do Internacional Int55511 junho 2024
Camiseta Feminina Baby Look do Internacional Int55511 junho 2024 -
 Rin-Nohara-online, rin nohara, naruto uzumaki11 junho 2024
Rin-Nohara-online, rin nohara, naruto uzumaki11 junho 2024 -
 Nextbots In Backrooms MOD APK 1.4.4 (No ads) Android11 junho 2024
Nextbots In Backrooms MOD APK 1.4.4 (No ads) Android11 junho 2024 -
 Minecraft: Story Mode APK For Android for Minecraft11 junho 2024
Minecraft: Story Mode APK For Android for Minecraft11 junho 2024 -
 COMO FOI A VOLTA DE MUTANTE REX NO BEN 10 REBOOT - BEN GEN 1011 junho 2024
COMO FOI A VOLTA DE MUTANTE REX NO BEN 10 REBOOT - BEN GEN 1011 junho 2024